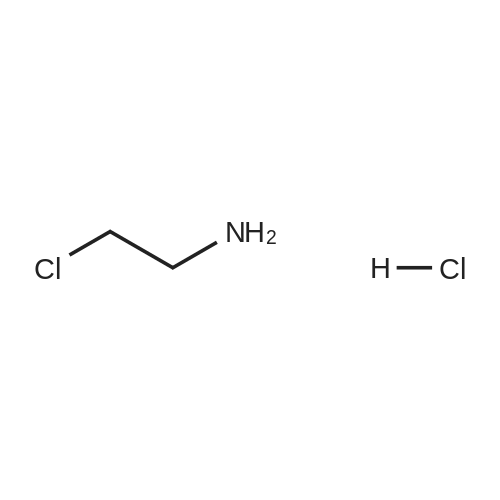


*Storage: {[sel_prStorage]}
*Shipping: {[sel_prShipping]}
4.5




 *For research use only!
*For research use only!
Change View
| Size | Price | VIP Price | USA Stock *0-1 Day |
Global Stock *5-7 Days |
In Stock | ||
| {[ item.pr_size ]} |
Inquiry
{[ getRatePrice(item.pr_usd, 1,1,item.pr_is_large_size_no_price, item.pr_usd) ]} {[ getRatePrice(item.pr_usd,item.pr_rate,1,item.pr_is_large_size_no_price, item.discount_usd) ]} {[ getRatePrice(item.pr_usd, 1,1,item.pr_is_large_size_no_price, item.pr_usd) ]} |
Inquiry {[ getRatePrice(item.pr_usd,item.pr_rate,item.mem_rate,item.pr_is_large_size_no_price, item.vip_usd) ]} | Inquiry {[ item.pr_usastock ]} In Stock Inquiry - | {[ item.pr_chinastock ]} {[ item.pr_remark ]} In Stock 1-2 weeks - Inquiry - | Login | - + | Inquiry |
Please Login or Create an Account to: See VIP prices and availability
1-2weeks
Inquiry
{[ getRatePrice(item.pr_usd,item.pr_rate,item.mem_rate,item.pr_is_large_size_no_price, item.vip_usd) ]}
{[ getRatePrice(item.pr_usd, 1,1,item.pr_is_large_size_no_price, item.pr_usd) ]}
{[ getRatePrice(item.pr_usd,1,item.mem_rate,item.pr_is_large_size_no_price, item.pr_usd) ]}
Inquiry
{[ getRatePrice(item.pr_usd,item.pr_rate,1,item.pr_is_large_size_no_price, item.vip_usd) ]}
{[ getRatePrice(item.pr_usd, 1,1,item.pr_is_large_size_no_price, item.pr_usd) ]}
{[ getRatePrice(item.pr_usd, 1,1,item.pr_is_large_size_no_price, item.pr_usd) ]}
In Stock
- +
Please Login or Create an Account to: See VIP prices and availability
Search for reports by entering the product batch number.
Batch number can be found on the product's label following the word 'Batch'.
Search for reports by entering the product batch number.
Batch number can be found on the product's label following the word 'Batch'.
Search for reports by entering the product batch number.
Batch number can be found on the product's label following the word 'Batch'.
Search for reports by entering the product batch number.
Batch number can be found on the product's label following the word 'Batch'.
Search for reports by entering the product batch number.
Batch number can be found on the product's label following the word 'Batch'.
| CAS No. : | 870-24-6 |
| Formula : | C2H7Cl2N |
| Linear Structure Formula : | ClCH2CH2NH2·HCl |
| M.W : | 115.99 |
| MDL No. : | MFCD00012887 |
| InChI Key : | ONRREFWJTRBDRA-UHFFFAOYSA-N |
| Pubchem ID : | 9793737 |
| GHS Pictogram: |

|
| Signal Word: | Warning |
| Hazard Statements: | H302-H315-H319-H335 |
| Precautionary Statements: | P261-P301+P312-P302+P352-P304+P340-P305+P351+P338 |
| Num. heavy atoms | 5 |
| Num. arom. heavy atoms | 0 |
| Fraction Csp3 | 1.0 |
| Num. rotatable bonds | 1 |
| Num. H-bond acceptors | 1.0 |
| Num. H-bond donors | 1.0 |
| Molar Refractivity | 26.2 |
| TPSA ? Topological Polar Surface Area: Calculated from |
26.02 ?2 |
| Log Po/w (iLOGP)? iLOGP: in-house physics-based method implemented from |
0.0 |
| Log Po/w (XLOGP3)? XLOGP3: Atomistic and knowledge-based method calculated by |
0.68 |
| Log Po/w (WLOGP)? WLOGP: Atomistic method implemented from |
0.99 |
| Log Po/w (MLOGP)? MLOGP: Topological method implemented from |
0.75 |
| Log Po/w (SILICOS-IT)? SILICOS-IT: Hybrid fragmental/topological method calculated by |
0.24 |
| Consensus Log Po/w? Consensus Log Po/w: Average of all five predictions |
0.53 |
| Log S (ESOL):? ESOL: Topological method implemented from |
-0.92 |
| Solubility | 13.9 mg/ml ; 0.12 mol/l |
| Class? Solubility class: Log S scale |
Very soluble |
| Log S (Ali)? Ali: Topological method implemented from |
-0.8 |
| Solubility | 18.2 mg/ml ; 0.157 mol/l |
| Class? Solubility class: Log S scale |
Very soluble |
| Log S (SILICOS-IT)? SILICOS-IT: Fragmental method calculated by |
-0.83 |
| Solubility | 17.3 mg/ml ; 0.149 mol/l |
| Class? Solubility class: Log S scale |
Soluble |
| GI absorption? Gatrointestinal absorption: according to the white of the BOILED-Egg |
High |
| BBB permeant? BBB permeation: according to the yolk of the BOILED-Egg |
Yes |
| P-gp substrate? P-glycoprotein substrate: SVM model built on 1033 molecules (training set) |
No |
| CYP1A2 inhibitor? Cytochrome P450 1A2 inhibitor: SVM model built on 9145 molecules (training set) |
No |
| CYP2C19 inhibitor? Cytochrome P450 2C19 inhibitor: SVM model built on 9272 molecules (training set) |
No |
| CYP2C9 inhibitor? Cytochrome P450 2C9 inhibitor: SVM model built on 5940 molecules (training set) |
No |
| CYP2D6 inhibitor? Cytochrome P450 2D6 inhibitor: SVM model built on 3664 molecules (training set) |
No |
| CYP3A4 inhibitor? Cytochrome P450 3A4 inhibitor: SVM model built on 7518 molecules (training set) |
No |
| Log Kp (skin permeation)? Skin permeation: QSPR model implemented from |
-6.52 cm/s |
| Lipinski? Lipinski (Pfizer) filter: implemented from |
0.0 |
| Ghose? Ghose filter: implemented from |
None |
| Veber? Veber (GSK) filter: implemented from |
0.0 |
| Egan? Egan (Pharmacia) filter: implemented from |
0.0 |
| Muegge? Muegge (Bayer) filter: implemented from |
3.0 |
| Bioavailability Score? Abbott Bioavailability Score: Probability of F > 10% in rat |
0.55 |
| PAINS? Pan Assay Interference Structures: implemented from |
0.0 alert |
| Brenk? Structural Alert: implemented from |
1.0 alert: heavy_metal |
| Leadlikeness? Leadlikeness: implemented from |
No; 1 violation:MW<1.0 |
| Synthetic accessibility? Synthetic accessibility score: from 1 (very easy) to 10 (very difficult) |
1.52 |
* All experimental methods are cited from the reference, please refer to the original source for details. We do not guarantee the accuracy of the content in the reference.

| Yield | Reaction Conditions | Operation in experiment |
|---|---|---|
| 56.9% | With sodium hydroxide In water at 70 - 75℃; for 4 h; | At first, water (5.Og) and aqueous phase from the methylation (5.Og, which containing approximately 0.0046 moles of the sodium methyl N-cyanodithioiminocarbonate, prepared as in Example 4 above) were taken in a 250 ml round bottom flask. The remaining aqueous phase from the methylation (71 .4g, containing approximately 0.066 moles of the sodium methyl N-cyanodithioiminocarbonate) and 70percent aqueous solution of 2- chloroethylammonium hydrochloride (20.0g, 70percent aqueous solution, 0.12 moles) were mixed in an addition funnel and this mixture was added to the flask over 4 hours at 70 —75°C maintaining the pH of the reaction mixture between 6.3 and 6.8 with concomitant addition of 50percent aqueous NaOH. The rest of the procedure was the same as in Example 4 above. Here, the weight of the dried CIT product was 6.9g with a purity of 97.8percent. The organic impurities as seen on LC were 0.9percent while the rest were inorganic salts, for example, sodium methyl sulfate and sodium chloride. The isolated yield of CIT based on cyanamide was 5 6.9percent. |
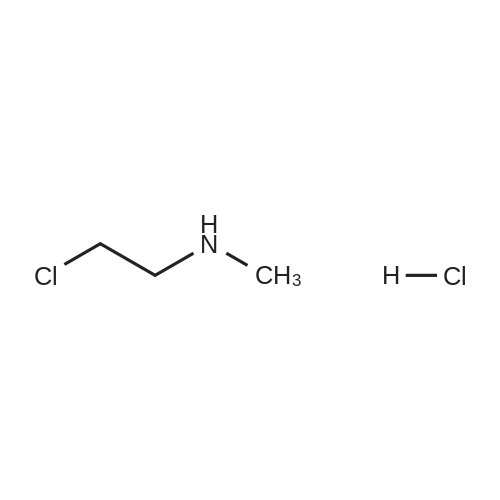
A102046[ 4535-90-4 ]
2-Chloro-N-methylethanamine hydrochloride
Similarity: 0.75
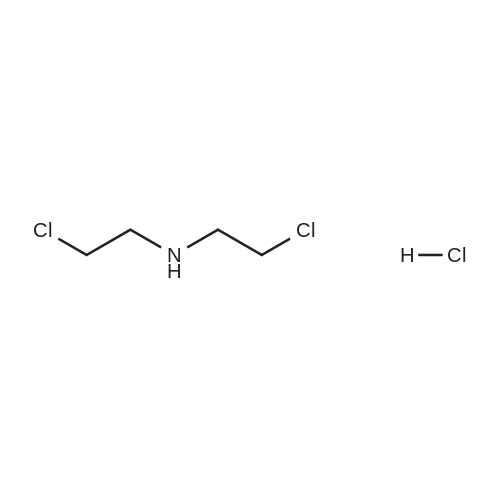
A104069[ 821-48-7 ]
Bis(2-Chloroethyl)amine hydrochloride
Similarity: 0.69
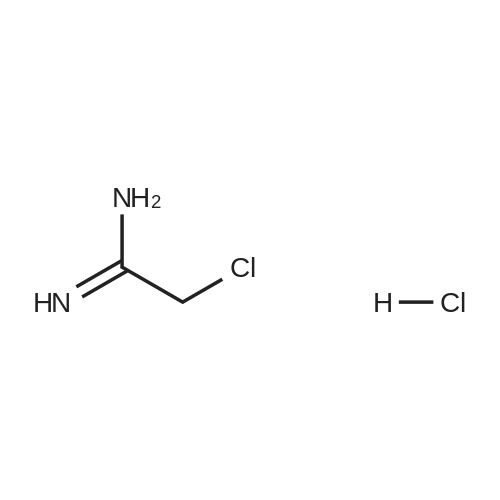
A186338[ 10300-69-3 ]
2-Chloroacetimidamide hydrochloride
Similarity: 0.64
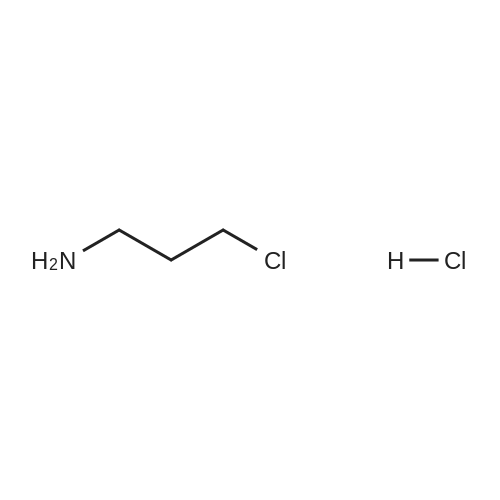
A257632[ 6276-54-6 ]
3-Chloropropan-1-amine hydrochloride
Similarity: 0.62

A102046[ 4535-90-4 ]
2-Chloro-N-methylethanamine hydrochloride
Similarity: 0.75

A104069[ 821-48-7 ]
Bis(2-Chloroethyl)amine hydrochloride
Similarity: 0.69

A186338[ 10300-69-3 ]
2-Chloroacetimidamide hydrochloride
Similarity: 0.64

A257632[ 6276-54-6 ]
3-Chloropropan-1-amine hydrochloride
Similarity: 0.62

A102046[ 4535-90-4 ]
2-Chloro-N-methylethanamine hydrochloride
Similarity: 0.75

A104069[ 821-48-7 ]
Bis(2-Chloroethyl)amine hydrochloride
Similarity: 0.69

A186338[ 10300-69-3 ]
2-Chloroacetimidamide hydrochloride
Similarity: 0.64

A257632[ 6276-54-6 ]
3-Chloropropan-1-amine hydrochloride
Similarity: 0.62